Principal Investigator
Associate Professor of Ophthalmology, Microbiology and Molecular Genetics, and Developmental Biology
University of Pittsburgh School of Medicine
Contact Information
BST 3-5060
3501 Fifth Avenue
Pittsburgh, PA 15213
412-383-5845
weix@upmc.edu
Research Interests
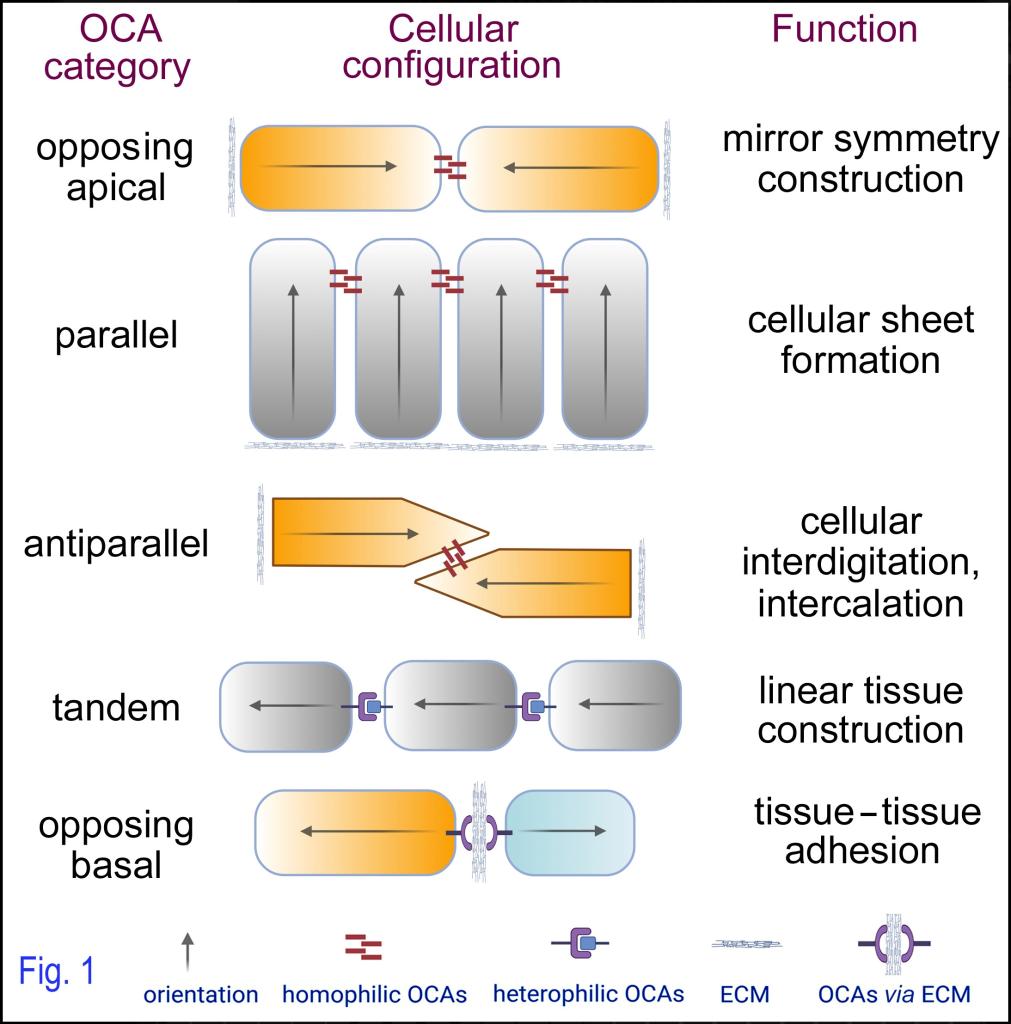 We are interested in how polarity genes regulate tissue morphogenesis and maintenance via cell adhesion. Based on our prior work, we recently proposed the concept of “orientational cell adhesions (OCAs)” to couple cell orientations with cell adhesions (Fig.1). OCAs refer to cell–cell, and cell–matrix–cell adhesions that define distinct orientational intercellular relationships, i.e., the relative orientations of the intrinsic polarities of coalesced cells. OCAs can be classified according to the distinct orientational intercellular relationships that they define, such as opposing apical, parallel, antiparallel, tandem, or opposing basal OCAs. OCAs complement conventional adhesions by emphasizing different aspects of cell adhesions.
We are interested in how polarity genes regulate tissue morphogenesis and maintenance via cell adhesion. Based on our prior work, we recently proposed the concept of “orientational cell adhesions (OCAs)” to couple cell orientations with cell adhesions (Fig.1). OCAs refer to cell–cell, and cell–matrix–cell adhesions that define distinct orientational intercellular relationships, i.e., the relative orientations of the intrinsic polarities of coalesced cells. OCAs can be classified according to the distinct orientational intercellular relationships that they define, such as opposing apical, parallel, antiparallel, tandem, or opposing basal OCAs. OCAs complement conventional adhesions by emphasizing different aspects of cell adhesions.
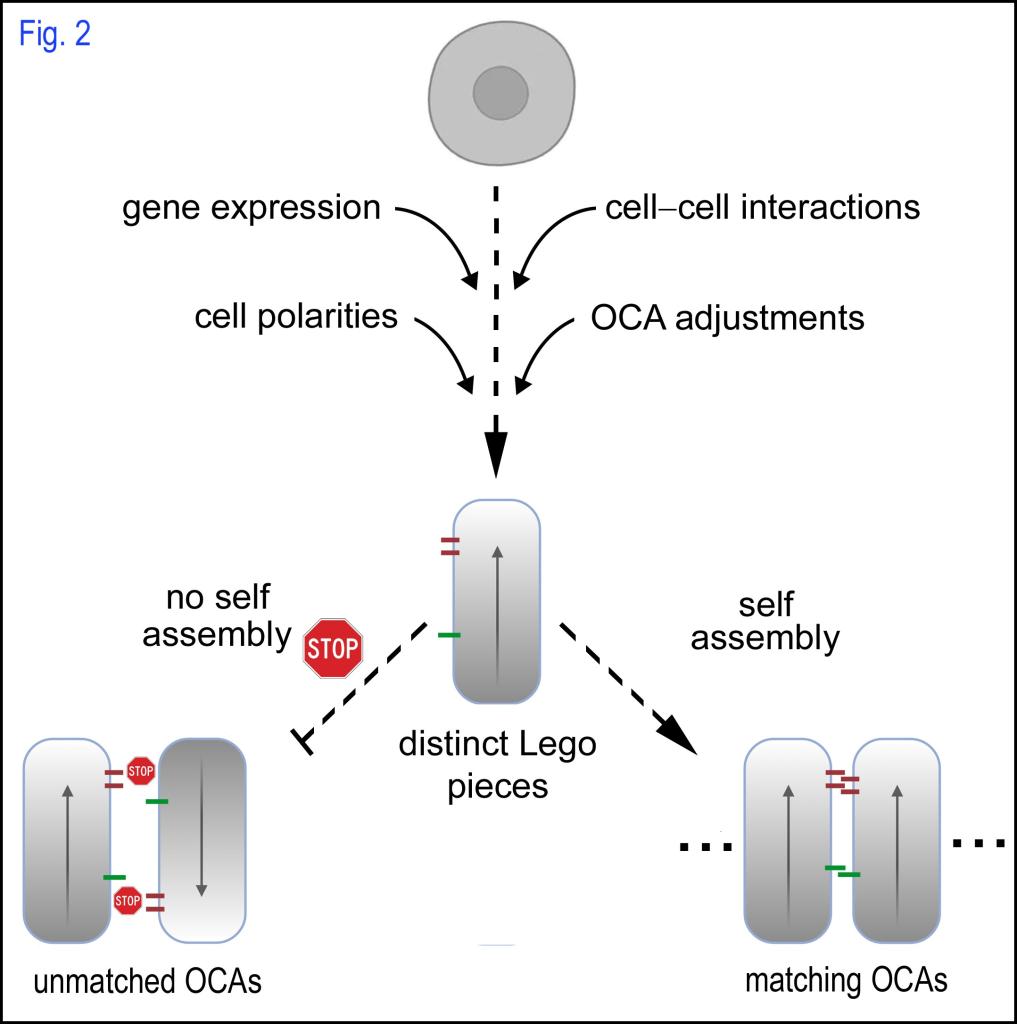
Stemming from the OCA concept, we further proposed “the Lego hypothesis of tissue morphogenesis”, which states that the topographical properties of cell surface adhesion molecules can be dynamically altered and polarized by regulating the spatiotemporal expression and localization of OCA molecules cell-autonomously and non-cell-autonomously, thus modulating cells into unique Lego pieces for self-assembling into distinct cytoarchitectures (Fig. 2). The Lego hypothesis demystifies tissue morphogenesis and simplifies it as a self-assembling process via OCAs.
The OCA concept and the Lego hypothesis offer a new perspective for us to study how cell adhesions regulate tissue morphogenesis and maintenance.
Research Directions
To better understand the molecular and cellular mechanisms by which OCAs regulate cellular pattern formation in tissue morphogenesis and maintenance, we are conducting research in four directions as described below.
(1) Synapse development and wiring
Cone and rod photoreceptors develop distinct synaptic terminals—pedicles and spherules—to wire properly with downstream neurons for normal vision. The mechanisms underlying their morphogenesis and connectivity are unclear. We study how polarity proteins regulate photoreceptor synaptogenesis and wiring through tandem OCAs. This research will provide insight into improving photoreceptor development and wiring in cell transplantation therapies, which offer hope for curing photoreceptor-based blindness.
(2) Neural tube closure
Neural tube closure defects (NTDs) lead to lethal anencephaly and debilitating spina bifida. The molecular basis for NTDs is not fully understood. We study how apical parallel, antiparallel, and opposing OCAs regulate neural tube closure and how the process is affected by genetic defects and adverse environmental factors. Our research will facilitate the development of new strategies for the diagnosis and prevention of NTDs.
(3) Retinal polarity gene transcription
Cell-type-specific expression of polarity genes is essential for proper cellular patterning of the neural retina. However, it is unclear how polarity genes are differentially expressed among various types of retinal cells. We study the regulation of polarity gene transcription at three levels: cis, trans, and nuclear spatial organization. The significance of this study goes beyond retinal morphogenesis and vision because polarity genes are essential for all tissues.
(4) Intervertebral disc homeostasis
Degenerated intervertebral discs (IVDs) can deform and press the surrounding spinal cord and nerves, thus causing back and neck pain, which affects two-thirds of people. The etiologies of IVD degeneration are not clear, and there is no cure. One critical tissue affected in degenerated IVDs is the nucleus pulposus (NP). We study how random OCAs regulate NP cellular organization and gene expression for NP maintenance. Our work has the potential to inspire new strategies for preventing and treating IVD degeneration.
Select Recent Publications
- Rainbow Enhancers Regulate Restrictive Transcription in Teleost Green, Red, and Blue Cones. Fang W, Guo C, Wei X. J Neurosci. 2017 Mar 15;37(11):2834-2848. doi: 10.1523/JNEUROSCI.3421-16.2017. Epub 2017 Feb 13.
- Crb apical polarity proteins maintain zebrafish retinal cone mosaics via intercellular binding of their extracellular domains. Zou J, Wang X, Wei X. Developmental Cell. 22:1261-1274. Epub 2012 May 10.
- Stepwise maturation of apicobasal polarity of the neuroepithelium is essential for vertebrate neurulation. Yang X, Zou J, Hyde D, Davidson L, Wei X. Journal of Neuroscience. 29:11426-11440. 2009 Sep 16;29(37):11426-40. doi: 10.1523/JNEUROSCI.1880-09.2009. Highlighted by a journal commentary: Premature Lin7c Expression Produces Multiaxial Mirror Symmetry. J. Neuroscience. This Week in the Journal, 29(37): i.i.
- Intact RPE maintained by Nok is essential for retinal epithelial polarity and cellular patterning in zebrafish. Zou J, Lathrop K, Sun M, Wei X. Journal of Neuroscience. 2008 Dec 10;28(50):13684-95. doi: 10.1523/JNEUROSCI.4333-08.2008.
- nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Wei X, Malicki J. Nature Genetics. 2002 Jun;31(2):150-7. doi: 10.1038/ng883. Epub 2002 May 6.
- Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. Wei X, Somanathan S, Samarabandu J, Berezney R. Journal of Cell Biology. 1999 Aug 9;146(3):543-58. doi: 10.1083/jcb.146.3.543. Highlighted by a journal commentary: Matrix-associated Transcription Sites in Three-dimensional Networks. Journal of Cell Biology “In Brief” 146 (3): 1.
- Segregation of transcription and replication sites into higher order domains. Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Science. 1998 Sep 4;281: 1502-1505. Highlighted by an Enhanced Perspective commentary by Dr. Peter Cook (Oxford University): Duplicating a Tangled Genome. Science. 281: 1466 – 1467. Also highlighted by another journal commentary: Taking turns at the genome. Science, “This Week in Science” 281:5382.

